Discovering cutting-edge functional genomics in Oklahoma
My research is focused on improving our understanding of the genetics of psoriasis; a skin condition that affects roughly 1.8 million people in the UK.
In the past couple of decades, researchers have identified hundreds of common genetic variants that increase risk of psoriasis. However, most of these common risk variants lie outside of coding regions of genes. This means most have an undefined function, and it is difficult to translate these findings into the clinic. What we do know is that risk variants are often found in important regions of DNA that regulate gene function, known as enhancers. These enhancers form physical looping interactions with their target genes.
It is important to characterise DNA looping between enhancers and target genes in disease, in order to to better understand disease pathways and identify new drugs targets. Without defining DNA looping we don’t know which genes are affected in each region, since enhancers can interact with distant, or even multiple genes.
Exploring new genomics techniques
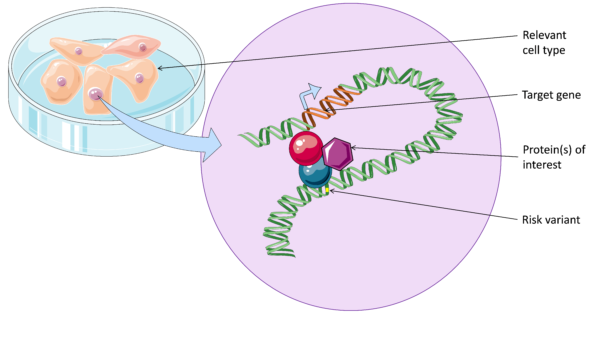
In 2016, a new genomics technique called HiChIP was developed at Stanford University (Mumbach et al., 2016). HiChIP is unique in that it combines information from two different aspects of genome regulation: (1) DNA looping and (2) protein binding. In short it allows us to detect long-range interactions that are associated with the binding of a specific protein of interest.
After hearing about the technique, I set out to use HiChIP to detect the interactions between enhancers and target genes in psoriasis. My plan is to carry out HiChIP for a protein marker of enhancers in psoriasis-relevant cells, such as skin cells and T lymphocytes.
However, still a relatively new technique, only a few teams have experience in using the process.
Through connections in the NIHR Manchester Biomedical Research Centre, I was able to arrange a lab visit to meet Dr Patrick Gaffney and his post-doc, Dr Yau Fu who are experienced in the HiChIP method, at the Oklahoma Medical Research Foundation (OMRF).
Their research focuses on characterising the genetics of lupus, and the team have published on a wide range of functional genomic techniques, including HiChIP. Accompanied by my colleague Dr Jenny Hankinson were excited to visit Oklahoma and observe the technique.
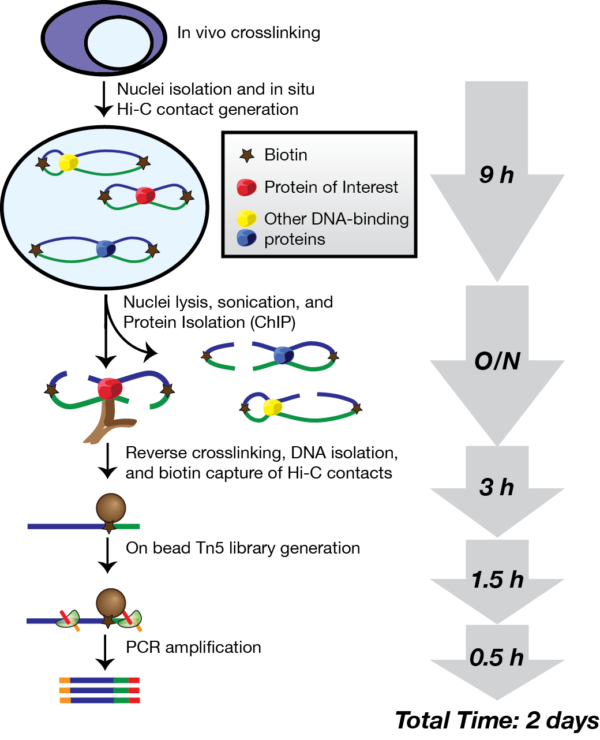
In the morning we headed straight to the lab at OMRF. Yao was planning to make HiChIP libraries using primary B cells from healthy donors. She had already performed crosslinking on the cells, which binds DNA and proteins together in their native conformation.
The HiChIP protocol takes about four days. First, Yao broke open the cells with a lysis buffer and cut up the DNA into fragments using a restriction enzyme. The ends of the fragments were then marked with biotin, which is used at a later point in the protocol to enrich for the DNA interactions. The ends of the fragments were then joined together overnight using DNA ligase. It was fascinating being able to watch and learn more about these initial processes.
Despite our jet lag and the heat, we were very happy to have a tour of the campus with Yao over lunch time. The campus, containing university and hospital buildings, was very beautiful and surprisingly lush. Apparently we had arrived in an unusually cool period, although it was still 30°C.
Wednesday started with Yao using a sonicator, a machine that uses energy to generate cavitation bubbles within the sample. When the bubbles collapse, high-energy jets of liquid are created. This was used to break open the nuclei and fragment the DNA into lengths of a suitable size for next-generation sequencing. An antibody was then added in order to pull out the regions of DNA binding a particular protein of interest: here a modified histone mark known as H3K27ac, which binds to active enhancers.
Today we also had a lab meeting. Dr Gaffney’s group are particularly interested in identifying causal genetic variants and underlying genetic mechanisms in lupus. Knowing which specific variants lead to disease becomes very important in the age of precision medicine. New therapeutic strategies, such as genome modification, require a very detailed understanding of the genetic signals underlying disease risk. Currently in lupus, as in most autoimmune diseases, our understanding is still far from complete.


Today our aim was to capture the antibody-bound DNA using special magnetic beads, and then remove the DNA from the beads and reversing the crosslinks using an enzyme called proteinase K.
We also had time to talk to some more of the other lab members. I had a discussion with a PhD student Jaanam Gopalakrishnan, who wants to use a technique known as 3C; something I used extensively during my PhD. Essentially it is a simplified version of HiChIP, but allows you to compare DNA looping intensities between samples of different genotypes. This is important if you want to see whether a disease-risk allele has an effect on DNA looping, compared with the protective allele.
It was great to be able to speak to other researchers and share good practice and knowledge. Science is all about collaboration.
This afternoon, Yao took us to the National Cowboy and Western Heritage Museum. Inside they had created a mock-up version of an old American frontier town. The sculptures and paintings were also incredible. I found out that women have a long history of competing in rodeo – an alternative career path maybe?
his was the most intensive day in the HiChIP process. Firstly the DNA was purified and quantified, after which the DNA interacting fragments marked by biotin (from day 1) were isolated using streptavidin, which binds very strongly to the biotin.
By now, the samples were enriched for (1) DNA contacts and (2) enhancer regions. The samples were prepared for next-generation sequencing using the transposase enzyme Tn5, which adds Illumina sequencing adaptors onto the fragment ends. After a final polymerase chain reaction, the sample quality was tested using a TapeStation, which shows the average fragment length (ideally around 200-700 base pairs) and concentration. Of course, Yao had succeeded in making made some high quality HiChIP libraries!
Being able to watch the process step by step, asking questions along the way was such an informative experience. I had found it frustrating not being able to complete the process in Manchester, but now I could see how it was performed and identify where I may have gone wrong.
This afternoon we were shown around the OMRF sequencing core, where the HiChIP libraries will be sequenced. We were introduced to the equipment by Dr Graham Wiley, who manages the facility. The core has an impressive array of sequencers, including the NovaSeq 6000, which sequences DNA using a new method known as “exclusion amplification”, and the MinION by Oxford Nanopore, which was invented here in the UK. The HiChIP libraries will be sequenced on the NextSeq 500 machine, generating around 20 million reads per library.


Last few days
As it was the weekend, Dr. Gaffney and his wife took us to a music concert at a historic Western town north of the city called Guthrie. The well-known fiddle player Bryon Berline put on an incredible performance of bluegrass alongside his band members who played the double bass, guitar, banjo and the “bones”. The band and the audience were quite excited to have two Brits among them, so we got an honourable mention and even a photo with the band at the end.
The remainder of our trip had passed by in a flash. It was really inspiring to visit Dr Gaffney’s lab and find out how they are using HiChIP to improve our understanding of autoimmune disease. I’m looking forward to further collaborative work between our labs, and excited to try the HiChIP technique back in Manchester!
Reference
Mumbach, M.R., Rubin, A.J., Flynn, R.A., Dai, C., Khavari, P.A., Greenleaf, W.J., and Chang, H.Y. (2016). HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat Methods 13, 919-922.
